Contamination aware approach: Vaiomer metagenomics is dedicated to low biomass samples
Metagenomics of low biomass samples is a challenge!
Blood and internal tissue microbiomes are more challenging to explore than the bacterial communities inhabiting body parts exposed to the outside environment such as the gut, the oral cavity or the skin. There are three major hurdles to overcome to analyze low bacterial biomass samples:
The low amount of bacterial DNA and more precisely the ratio between bacterial and host DNA. In high bacterial biomass samples, the amount of bacterial DNA is highly superior to the amount of host DNA. The microbial-to-host DNA ratio is critically lowered with blood and tissue microbiomes.
The high amount of PCR inhibitors in blood and other internal organs makes the DNA of these bacterial communities more difficult to amplify.
The considerable susceptibility to potential contamination from the environment (operating room, dissection, sample storage and processing), reagents and consumables could lead to false-positive results.
Vaiomer unique technology and contamination-aware approach
Vaiomer successfully overcame these obstacles to investigate low biomass samples by developing and optimizing a tissue-specific metagenomic approach combining:
Tissue-specific DNA extraction to maximize the recovery of bacterial DNA and to minimize DNA contamination.
Quantitative PCR (qPCR) targeting the V3-V4 variable regions of the 16S rRNA gene. Vaiomer optimized qPCR is highly sensitive and reliable to quantify low to very low amount of microbial DNA. It is also a powerful guide to exclude the potential impact of contamination.
16S rRNA gene sequencing with the amplification step targeting the same V3-V4 variable regions of the bacterial 16S rRNA gene. The sequencing is performed on the Illumina MiSeq system and the data is analyzed on a tailor-made bioinformatics workflow. This technique is used to identify the bacterial component of the microbiome.
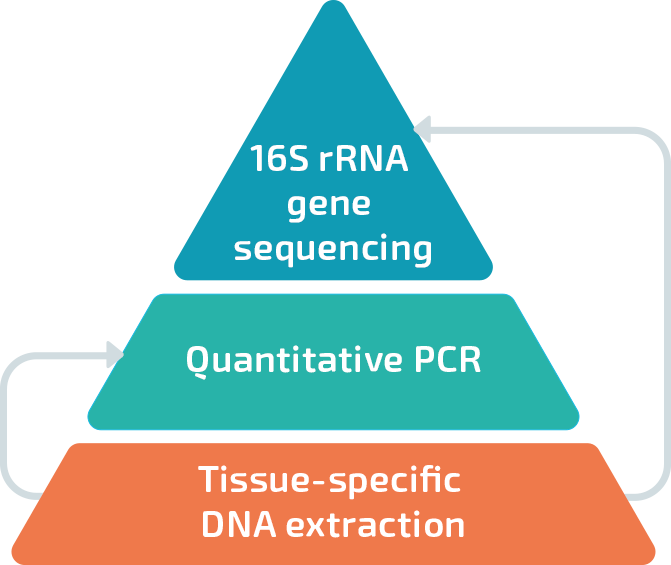
Vaiomer methods are developed to work on high to very low bacterial biomass. The amplifications of the bacterial 16S rRNA gene are optimized for low amount of bacterial DNA and a wide range of PCR inhibitors. Because both the qPCR and the sequencing target the same part of the 16S rDNA, the quantification and the taxonomic profile of the microbiomes are easily integrable.
The presence of bacteria and bacterial products circulating inside the body and their depositing on different internal tissues is subject of intense debate. Vaiomer contamination-aware approach accounts for potential DNA contamination by deploying specific countermeasures and by using a comprehensive set of negative controls to manage the risk of reporting false-positive results.
Microbiome analysis: quantification and taxonomic profiling
Vaiomer technology couples quantitative PCR and targeted metagenomics to characterize high to very low bacterial biomass samples. The quantification of the bacterial DNA load in blood and tissues allows to indirectly assess bacterial translocation and thus inflammation.
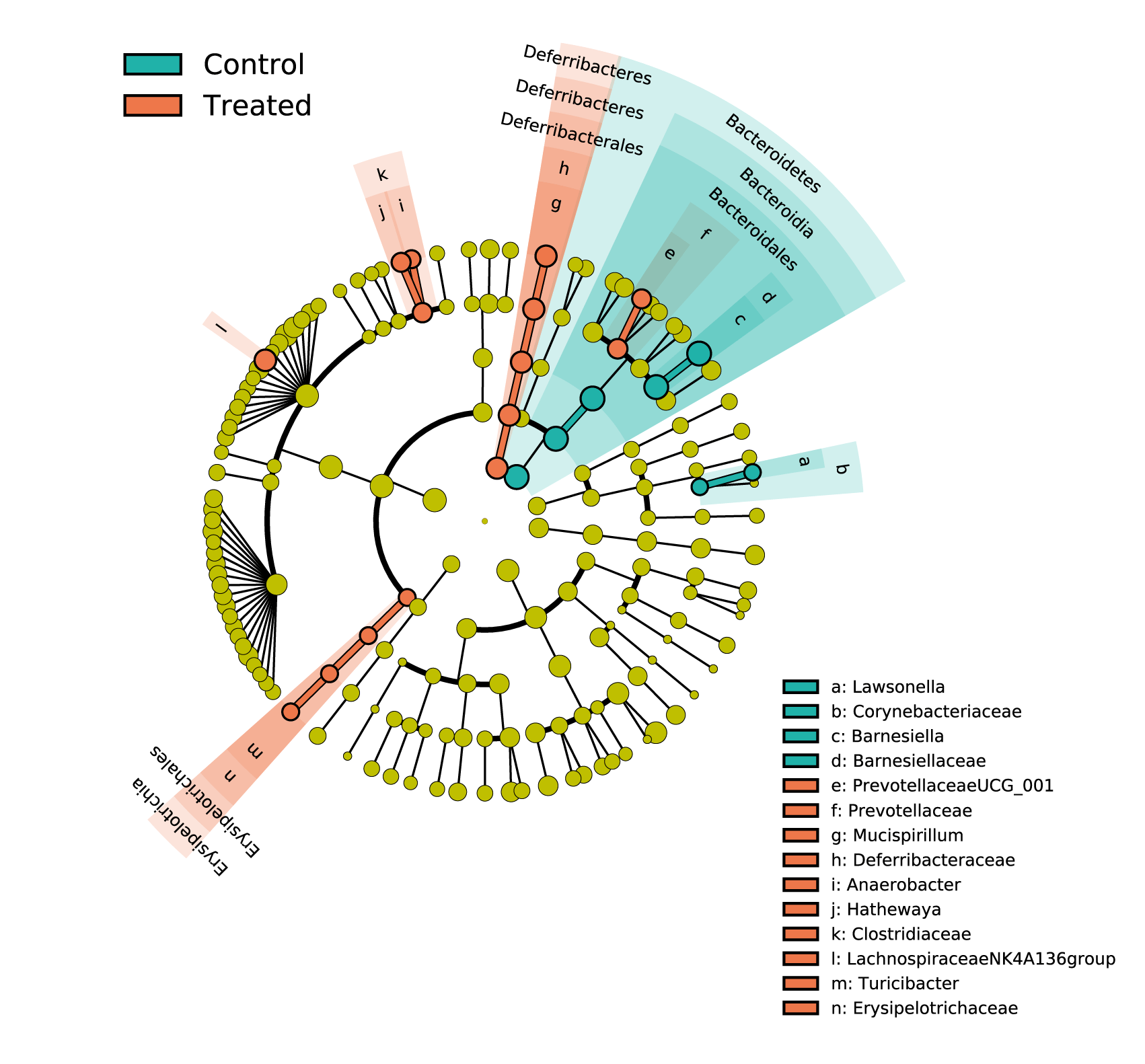
The 16S rRNA gene sequencing allows the profiling of the bacterial communities. From high quality reads, sequences are clusterized into operational taxonomic units (OTUs) for diversity analysis and taxonomic assignation. Samples are compared on their measurements of alpha-diversity and beta-diversity as well as on the composition and relative abundance of their microbiome. The 16S rDNA sequencing profiles can also be used to predict the metabolic potential of the microbiome thanks to PICRUSt2. The predicted bacterial genes and pathways can give insights into functional microbial ecology.
Customized statistical analyses are also implemented for group comparisons to highlight significant differences. Non-parametric and parametric testing are applied on the quantification data. Linear Discriminant Analysis Effect Size (LEfSe) is performed to characterize the tissue-specific taxonomic features that best discriminate biological groups.
Planning to start a microbiota study? See how Vaiomer can help you!
Q&A about microbiome studies in low biomass samples
Why sequence the bacterial 16S rRNA gene?
The 16S rRNA gene is shared among all bacteria. It contains conserved regions allowing its amplification and variable regions whose diversity can be used to discriminate and identify bacterial taxa.
Why use 16S rRNA gene sequencing and not whole genome sequencing ?
To this date, due to technical limitations, 16S rRNA gene sequencing is the best metagenomic approach for low bacterial biomass samples. In these types of samples, whole genome sequencing only yields sequences that belong to the host.
Why target the V3-V4 variable regions of the bacterial 16S rRNA gene?
What type of samples can be analyzed?
Look at our atlas of microbiomes for a non-exhaustive list of samples that have already been analyzed with Vaiomer technology. We are always looking forward to new challenges, so please contact us with your specific needs.
How much tissue material do I need?
Vaiomer metagenomics is highly sensitive so only a very small amount of material is needed and compatible with common clinical and animal experimentation procedures.
How should I store my samples?
Your samples should ideally be stored dried (without any solution) and immediately put at -80°C.
What are the sources of contamination in microbiome studies?
Isn’t blood supposed to be sterile?
Many reliable studies have now shown that the blood is not sterile even in healthy individuals. The old dogma of sterile blood was never demonstrated. Please see the microbiome discovery timeline to learn more about the history of bacteria in the blood.
What does contamination-aware mean?
Vaiomer approach does not overlook potential contaminants and their sources but rather deploys specific countermeasure to reduce them to their minimum. Vaiomer also uses comprehensive sets of negative controls to confirm that potential DNA contaminants have little to no impact on the biological samples.
What sequence analysis method do you use?
We use SWARM, a modern approach for OTU clustering that does not use arbitrary global clustering thresholds of similarity but rather creates robust and more natural OTUs.
Do you perform the bioinformatics and statistical analyses?
We discuss with you before the start of your study to define the best analysis plan for your data and perform the bioinformatics and statistics accordingly.
What type of data do you deliver?
When your study is completed, we send you a report of your study including figures and tables to illustrate your results. We also deliver you the raw data and other intermediate files produced during the bioinformatic analysis.
What are the applications of microbiome studies?
INNOVATORS’ EXPERIENCE
The early explorers of low biomass microbiomes









More details about metagenomics and the control of contaminants?

Join Benjamin for a first chat.
Benjamin LELOUVIER
Chief Scientific Officer